pylops.Spread¶
-
class
pylops.Spread(dims, dimsd, table=None, dtable=None, fh=None, interp=False, engine='numpy', dtype='float64')[source]¶ Spread operator.
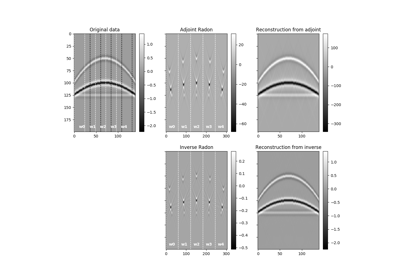
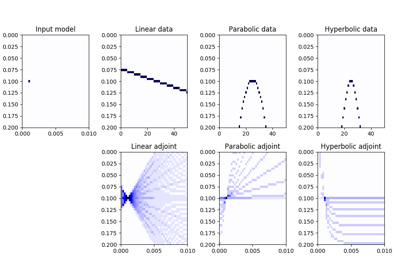
Spread values from the input model vector arranged as a 2-dimensional array of size \([n_{x0} \times n_{t0}]\) into the data vector of size \([n_x \times n_t]\). Spreading is performed along parametric curves provided as look-up table of pre-computed indices (
table) or computed on-the-fly using a function handle (fh).In adjont mode, values from the data vector are instead stacked along the same parametric curves.
Parameters: - dims :
tuple Dimensions of model vector (vector will be reshaped internally into a two-dimensional array of size \([n_{x0} \times n_{t0}]\), where the first dimension is the spreading/stacking direction)
- dimsd :
tuple Dimensions of model vector (vector will be reshaped internal into a two-dimensional array of size \([n_x \times n_t]\))
- table :
np.ndarray, optional Look-up table of indeces of size \([n_{x0} \times n_{t0} \times n_x]\) (if
Noneuse function handlefh)- dtable :
np.ndarray, optional Look-up table of decimals remainders for linear interpolation of size \([n_{x0} \times n_{t0} \times n_x]\) (if
Noneuse function handlefh)- fh :
np.ndarray, optional Function handle that returns an index (and a fractional value in case of
interp=True) to be used for spreading/stacking given indices in \(x0\) and \(t\) axes (ifNoneuse look-up tabletable)- interp :
bool, optional Apply linear interpolation (
True) or nearest interpolation (False) during stacking/spreading along parametric curve. To be used only ifengine='numba', inferred directly from the number of outputs offhforengine='numpy'- engine :
str, optional Engine used for fft computation (
numpyornumba). Note thatnumbacan only be used when providing a look-up table- dtype :
str, optional Type of elements in input array.
Raises: - KeyError
If
engineis neithernumpynornumba- NotImplementedError
If both
tableandfhare not provided- ValueError
If
tablehas shape different from \([n_{x0} \times n_t0 \times n_x]\)
Notes
The Spread operator applies the following linear transform in forward mode to the model vector after reshaping it into a 2-dimensional array of size \([n_x \times n_t]\):
\[m(x0, t_0) \rightarrow d(x, t=f(x0, x, t_0))\]where \(f(x0, x, t)\) is a mapping function that returns a value t given values \(x0\), \(x\), and \(t_0\).
In adjoint mode, the model is reconstructed by means of the following stacking operation:
\[m(x0, t_0) = \int{d(x, t=f(x0, x, t_0))} dx\]Note that
table(orfh) must return integer numbers representing indices in the axis \(t\). However it also possible to perform linear interpolation as part of the spreading/stacking process by providing the decimal part of the mapping function (\(t - \lfloor t \rfloor\)) either indtableinput parameter or as second value in the return offhfunction.Attributes: Methods
__init__(self, dims, dimsd[, table, dtable, …])Initialize this LinearOperator. adjoint(self)Hermitian adjoint. apply_columns(self, cols)Apply subset of columns of operator cond(self, \*\*kwargs_eig)Condition number of linear operator. conj(self)Complex conjugate operator div(self, y[, niter])Solve the linear problem \(\mathbf{y}=\mathbf{A}\mathbf{x}\). dot(self, x)Matrix-matrix or matrix-vector multiplication. eigs(self[, neigs, symmetric, niter])Most significant eigenvalues of linear operator. matmat(self, X)Matrix-matrix multiplication. matvec(self, x)Matrix-vector multiplication. rmatmat(self, X)Adjoint matrix-matrix multiplication. rmatvec(self, x)Adjoint matrix-vector multiplication. todense(self)Return dense matrix. transpose(self)Transpose this linear operator. - dims :