Note
Click here to download the full example code
OMP, ISTA and FISTA¶
This example shows how to use the pylops.optimization.sparsity.OMP,
pylops.optimization.sparsity.ISTA, and
pylops.optimization.sparsity.FISTA solvers.
These solvers can be used when the model to retrieve is supposed to have a sparse representation in a certain domain. OMP uses a L0 norm and mathematically translates to solving the following constrained problem:
while ISTA and FISTA solve an uncostrained problem with a L1 regularization term:
import numpy as np
import matplotlib.pyplot as plt
import pylops
plt.close('all')
np.random.seed(0)
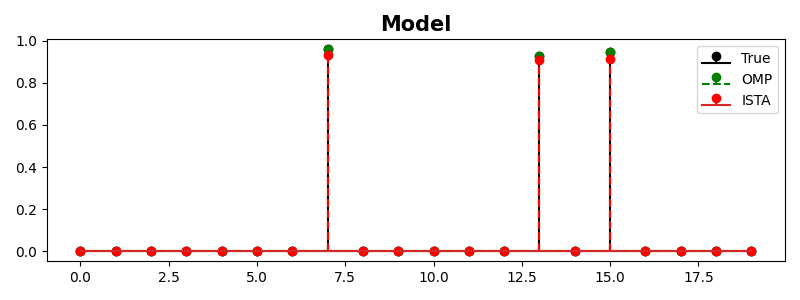
Let’s start with a simple example, where we create a dense mixing matrix and a sparse signal and we use OMP and ISTA to recover such a signal. Note that the mixing matrix leads to an underdetermined system of equations (\(N < M\)) so being able to add some extra prior information regarding the sparsity of our desired model is essential to be able to invert such a system.
N, M = 15, 20
Aop = pylops.MatrixMult(np.random.randn(N, M))
x = np.random.rand(M)
x[x < 0.9] = 0
y = Aop*x
# ISTA
eps = 0.5
maxit = 1000
x_omp = pylops.optimization.sparsity.OMP(Aop, y, maxit, sigma=1e-4)[0]
x_ista = pylops.optimization.sparsity.ISTA(Aop, y, maxit, eps=eps,
tol=0, returninfo=True)[0]
fig, ax = plt.subplots(1, 1, figsize=(8, 3))
ax.stem(x, linefmt='k', basefmt='k',
markerfmt='ko', label='True')
ax.stem(x_omp, linefmt='--g', basefmt='--g',
markerfmt='go', label='OMP')
ax.stem(x_ista, linefmt='--r',
markerfmt='ro', label='ISTA')
ax.set_title('Model', size=15, fontweight='bold')
ax.legend()
plt.tight_layout()
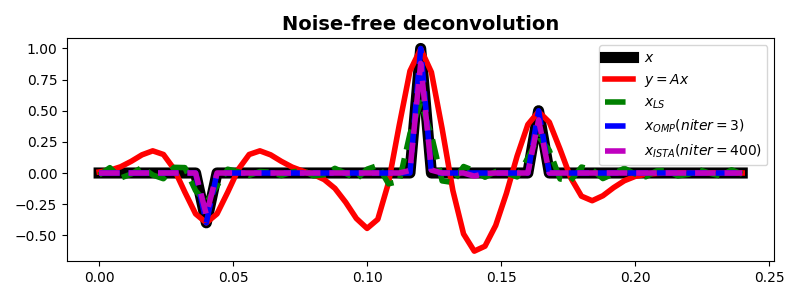
We now consider a more interesting problem problem, wavelet deconvolution
from a signal that we assume being composed by a train of spikes convolved
with a certain wavelet. We will see how solving such a problem with a
least-squares solver such as
pylops.optimization.leastsquares.RegularizedInversion does not
produce the expected results (especially in the presence of noisy data),
conversely using the pylops.optimization.sparsity.ISTA and
pylops.optimization.sparsity.FISTA solvers allows us
to succesfully retrieve the input signal even in the presence of noise.
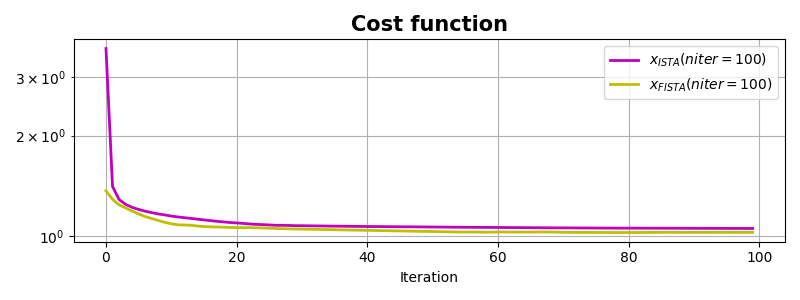
pylops.optimization.sparsity.FISTA shows faster convergence which
is particularly useful for this problem.
nt = 61
dt = 0.004
t = np.arange(nt)*dt
x = np.zeros(nt)
x[10] = -.4
x[int(nt/2)] = 1
x[nt-20] = 0.5
h, th, hcenter = pylops.utils.wavelets.ricker(t[:101], f0=20)
Cop = pylops.signalprocessing.Convolve1D(nt, h=h, offset=hcenter,
dtype='float32')
y = Cop*x
yn = y + np.random.normal(0, 0.1, y.shape)
# noise free
xls = Cop / y
xomp, nitero, costo = \
pylops.optimization.sparsity.OMP(Cop, y, niter_outer=200, sigma=1e-8)
xista, niteri, costi = \
pylops.optimization.sparsity.ISTA(Cop, y, niter=400, eps=5e-1,
tol=1e-8, returninfo=True)
fig, ax = plt.subplots(1, 1, figsize=(8, 3))
ax.plot(t, x, 'k', lw=8, label=r'$x$')
ax.plot(t, y, 'r', lw=4, label=r'$y=Ax$')
ax.plot(t, xls, '--g', lw=4, label=r'$x_{LS}$')
ax.plot(t, xomp, '--b', lw=4, label=r'$x_{OMP} (niter=%d)$' % nitero)
ax.plot(t, xista, '--m', lw=4, label=r'$x_{ISTA} (niter=%d)$' % niteri)
ax.set_title('Noise-free deconvolution', fontsize=14, fontweight='bold')
ax.legend()
plt.tight_layout()
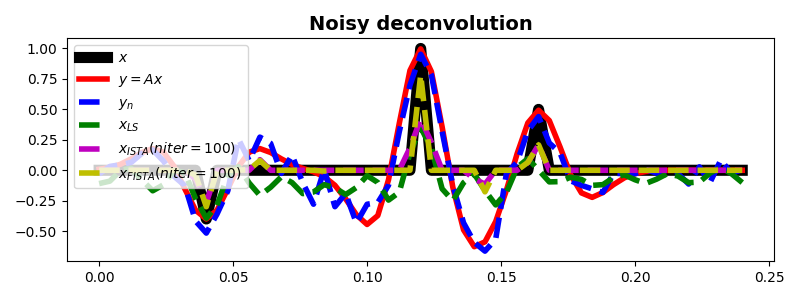
# noisy
xls = \
pylops.optimization.leastsquares.RegularizedInversion(Cop, [], yn,
returninfo=False,
**dict(damp=1e-1,
atol=1e-3,
iter_lim=100,
show=0))
xista, niteri, costi = \
pylops.optimization.sparsity.ISTA(Cop, yn, niter=100, eps=5e-1,
tol=1e-5, returninfo=True)
xfista, niterf, costf = \
pylops.optimization.sparsity.FISTA(Cop, yn, niter=100, eps=5e-1,
tol=1e-5, returninfo=True)
fig, ax = plt.subplots(1, 1, figsize=(8, 3))
ax.plot(t, x, 'k', lw=8, label=r'$x$')
ax.plot(t, y, 'r', lw=4, label=r'$y=Ax$')
ax.plot(t, yn, '--b', lw=4, label=r'$y_n$')
ax.plot(t, xls, '--g', lw=4, label=r'$x_{LS}$')
ax.plot(t, xista, '--m', lw=4, label=r'$x_{ISTA} (niter=%d)$' % niteri)
ax.plot(t, xfista, '--y', lw=4, label=r'$x_{FISTA} (niter=%d)$' % niterf)
ax.set_title('Noisy deconvolution', fontsize=14, fontweight='bold')
ax.legend()
plt.tight_layout()
fig, ax = plt.subplots(1, 1, figsize=(8, 3))
ax.semilogy(costi, 'm', lw=2, label=r'$x_{ISTA} (niter=%d)$' % niteri)
ax.semilogy(costf, 'y', lw=2, label=r'$x_{FISTA} (niter=%d)$' % niterf)
ax.set_title('Cost function', size=15, fontweight='bold')
ax.set_xlabel('Iteration')
ax.legend()
ax.grid(True, which='both')
plt.tight_layout()
Total running time of the script: ( 0 minutes 1.070 seconds)