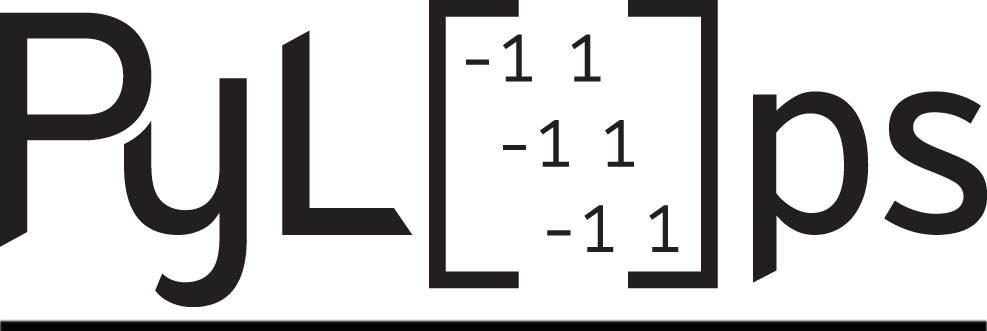pylops.waveeqprocessing.MDD#
- pylops.waveeqprocessing.MDD(G, d, dt=0.004, dr=1.0, nfmax=None, wav=None, twosided=True, causality_precond=False, adjoint=False, psf=False, dottest=False, saveGt=True, add_negative=True, smooth_precond=0, fftengine='numpy', **kwargs_solver)[source]#
Multi-dimensional deconvolution.
Solve multi-dimensional deconvolution problem using
scipy.sparse.linalg.lsqriterative solver.- Parameters
- G
numpy.ndarray Multi-dimensional convolution kernel in time domain of size \([n_s \times n_r \times n_t]\) for
twosided=Falseortwosided=Trueandadd_negative=True(with only positive times) or size \([n_s \times n_r \times 2n_t-1]\) fortwosided=Trueandadd_negative=False(with both positive and negative times)- d
numpy.ndarray Data in time domain \([n_s \,(\times n_{vs}) \times n_t]\) if
twosided=Falseortwosided=Trueandadd_negative=True(with only positive times) or size \([n_s \,(\times n_{vs}) \times 2n_t-1]\) iftwosided=True- dt
float, optional Sampling of time integration axis
- dr
float, optional Sampling of receiver integration axis
- nfmax
int, optional Index of max frequency to include in deconvolution process
- wav
numpy.ndarray, optional Wavelet to convolve to the inverted model and psf (must be centered around its index in the middle of the array). If
None, the outputs of the inversion are returned directly.- twosided
bool, optional MDC operator and data both negative and positive time (
True) or only positive (False)- add_negative
bool, optional Add negative side to MDC operator and data (
True) or not (False)- operator and data are already provided with both positive and negative sides. To be used only withtwosided=True.- causality_precond
bool, optional Apply causality mask (
True) or not (False)- smooth_precond
int, optional Lenght of smoothing to apply to causality preconditioner
- adjoint
bool, optional Compute and return adjoint(s)
- psf
bool, optional Compute and return Point Spread Function (PSF) and its inverse
- dottest
bool, optional Apply dot-test
- saveGt
bool, optional Save
GandG.Hto speed up the computation of adjoint ofpylops.signalprocessing.Fredholm1(True) or createG.Hon-the-fly (False) Note thatsaveGt=Truewill be faster but double the amount of required memory- fftengine
str, optional Engine used for fft computation (
numpy,scipyorfftw)- **kwargs_solver
Arbitrary keyword arguments for chosen solver (
scipy.sparse.linalg.cgandpylops.optimization.solver.cgare used as default for numpy and cupy data, respectively)
- G
- Returns
- minv
numpy.ndarray Inverted model of size \([n_r \,(\times n_{vs}) \times n_t]\) for
twosided=Falseor \([n_r \,(\times n_vs) \times 2n_t-1]\) fortwosided=True- madj
numpy.ndarray Adjoint model of size \([n_r \,(\times n_{vs}) \times n_t]\) for
twosided=Falseor \([n_r \,(\times n_r) \times 2n_t-1]\) fortwosided=True- psfinv
numpy.ndarray Inverted psf of size \([n_r \times n_r \times n_t]\) for
twosided=Falseor \([n_r \times n_r \times 2n_t-1]\) fortwosided=True- psfadj
numpy.ndarray Adjoint psf of size \([n_r \times n_r \times n_t]\) for
twosided=Falseor \([n_r \times n_r \times 2n_t-1]\) fortwosided=True
- minv
See also
MDCMulti-dimensional convolution
Notes
Multi-dimensional deconvolution (MDD) is a mathematical ill-solved problem, well-known in the image processing and geophysical community [1].
MDD aims at removing the effects of a Multi-dimensional Convolution (MDC) kernel or the so-called blurring operator or point-spread function (PSF) from a given data. It can be written as
\[\mathbf{d}= \mathbf{D} \mathbf{m}\]or, equivalently, by means of its normal equation
\[\mathbf{m}= (\mathbf{D}^H\mathbf{D})^{-1} \mathbf{D}^H\mathbf{d}\]where \(\mathbf{D}^H\mathbf{D}\) is the PSF.
- 1
Wapenaar, K., van der Neut, J., Ruigrok, E., Draganov, D., Hunziker, J., Slob, E., Thorbecke, J., and Snieder, R., “Seismic interferometry by crosscorrelation and by multi-dimensional deconvolution: a systematic comparison”, Geophyscial Journal International, vol. 185, pp. 1335-1364. 2011.