Note
Go to the end to download the full example code
Polynomial Regression#
This example shows how to use the pylops.Regression operator
to perform Polynomial regression analysis.
In short, polynomial regression is the problem of finding the best fitting coefficients for the following equation:
\[y_i = \sum_{n=0}^\text{order} x_n t_i^n \qquad \forall i=0,1,\ldots,N-1\]
As we can express this problem in a matrix form:
\[\mathbf{y}= \mathbf{A} \mathbf{x}\]
our solution can be obtained by solving the following optimization problem:
\[J= ||\mathbf{y} - \mathbf{A} \mathbf{x}||_2\]
See documentation of pylops.Regression for more detailed
definition of the forward problem.
import matplotlib.pyplot as plt
import numpy as np
import pylops
plt.close("all")
np.random.seed(10)
Define the input parameters: number of samples along the t-axis (N),
order (order), regression coefficients (x), and standard deviation
of noise to be added to data (sigma).
Let’s create the time axis and initialize the
pylops.Regression operator
We can then apply the operator in forward mode to compute our data points
along the x-axis (y). We will also generate some random gaussian noise
and create a noisy version of the data (yn).
We are now ready to solve our problem. As we are using an operator from the
pylops.LinearOperator family, we can simply use /,
which in this case will solve the system by means of an iterative solver
(i.e., scipy.sparse.linalg.lsqr).
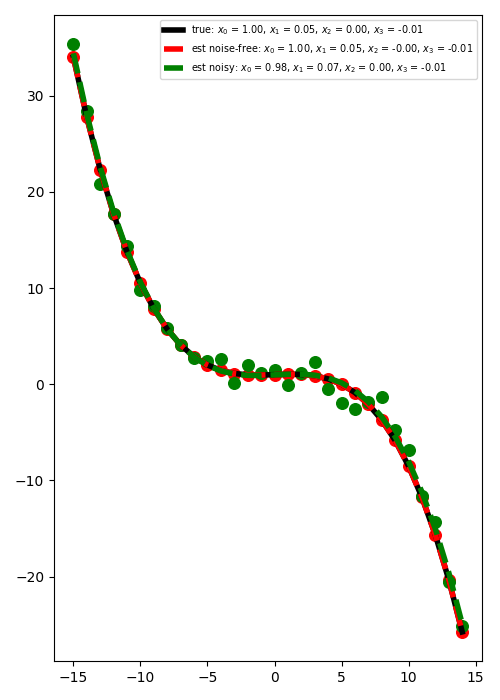
Let’s plot the best fitting curve for the case of noise free and noisy data
plt.figure(figsize=(5, 7))
plt.plot(
t,
PRop * x,
"k",
lw=4,
label=r"true: $x_0$ = %.2f, $x_1$ = %.2f, "
r"$x_2$ = %.2f, $x_3$ = %.2f" % (x[0], x[1], x[2], x[3]),
)
plt.plot(
t,
PRop * xest,
"--r",
lw=4,
label="est noise-free: $x_0$ = %.2f, $x_1$ = %.2f, "
r"$x_2$ = %.2f, $x_3$ = %.2f" % (xest[0], xest[1], xest[2], xest[3]),
)
plt.plot(
t,
PRop * xnest,
"--g",
lw=4,
label="est noisy: $x_0$ = %.2f, $x_1$ = %.2f, "
r"$x_2$ = %.2f, $x_3$ = %.2f" % (xnest[0], xnest[1], xnest[2], xnest[3]),
)
plt.scatter(t, y, c="r", s=70)
plt.scatter(t, yn, c="g", s=70)
plt.legend(fontsize="x-small")
plt.tight_layout()
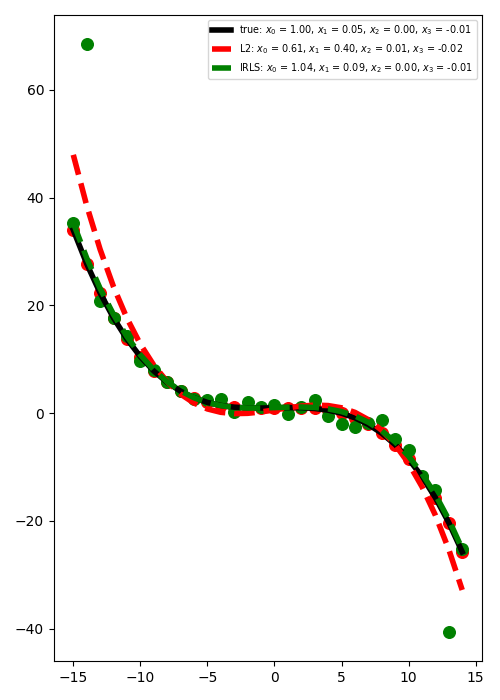
We consider now the case where some of the observations have large errors.
Such elements are generally referred to as outliers and can affect the
quality of the least-squares solution if not treated with care. In this
example we will see how using a L1 solver such as
pylops.optimization.sparsity.IRLS can drammatically improve the
quality of the estimation of intercept and gradient.
# Add outliers
yn[1] += 40
yn[N - 2] -= 20
# IRLS
nouter = 20
epsR = 1e-2
epsI = 0
tolIRLS = 1e-2
xnest = PRop / yn
xirls, nouter = pylops.optimization.sparsity.irls(
PRop,
yn,
nouter=nouter,
threshR=False,
epsR=epsR,
epsI=epsI,
tolIRLS=tolIRLS,
)
print(f"IRLS converged at {nouter} iterations...")
plt.figure(figsize=(5, 7))
plt.plot(
t,
PRop * x,
"k",
lw=4,
label=r"true: $x_0$ = %.2f, $x_1$ = %.2f, "
r"$x_2$ = %.2f, $x_3$ = %.2f" % (x[0], x[1], x[2], x[3]),
)
plt.plot(
t,
PRop * xnest,
"--r",
lw=4,
label=r"L2: $x_0$ = %.2f, $x_1$ = %.2f, "
r"$x_2$ = %.2f, $x_3$ = %.2f" % (xnest[0], xnest[1], xnest[2], xnest[3]),
)
plt.plot(
t,
PRop * xirls,
"--g",
lw=4,
label=r"IRLS: $x_0$ = %.2f, $x_1$ = %.2f, "
r"$x_2$ = %.2f, $x_3$ = %.2f" % (xirls[0], xirls[1], xirls[2], xirls[3]),
)
plt.scatter(t, y, c="r", s=70)
plt.scatter(t, yn, c="g", s=70)
plt.legend(fontsize="x-small")
plt.tight_layout()
IRLS converged at 6 iterations...
Total running time of the script: (0 minutes 0.574 seconds)