Note
Go to the end to download the full example code
Restriction and Interpolation#
This example shows how to use the pylops.Restriction operator
to sample a certain input vector at desired locations iava. Moreover,
we go one step further and use the pylops.signalprocessing.Interp
operator to show how we can also sample values at locations that are not
exactly on the grid of the input vector.
As explained in the 03. Solvers tutorial, such operators can be used as forward model in an inverse problem aimed at interpolate irregularly sampled 1d or 2d signals onto a regular grid.
import matplotlib.pyplot as plt
import numpy as np
import pylops
plt.close("all")
np.random.seed(10)
Let’s create a signal of size nt and sampling dt that is composed
of three sinusoids at frequencies freqs.
First of all, we subsample the signal at random locations and we retain 40% of the initial samples.
perc_subsampling = 0.4
ntsub = int(np.round(nt * perc_subsampling))
isample = np.arange(nt)
iava = np.sort(np.random.permutation(np.arange(nt))[:ntsub])
We then create the restriction and interpolation operators and display the original signal as well as the subsampled signal.
Rop = pylops.Restriction(nt, iava, dtype="float64")
NNop, iavann = pylops.signalprocessing.Interp(
nt, iava + 0.4, kind="nearest", dtype="float64"
)
LIop, iavali = pylops.signalprocessing.Interp(
nt, iava + 0.4, kind="linear", dtype="float64"
)
SIop, iavasi = pylops.signalprocessing.Interp(
nt, iava + 0.4, kind="sinc", dtype="float64"
)
y = Rop * x
ynn = NNop * x
yli = LIop * x
ysi = SIop * x
ymask = Rop.mask(x)
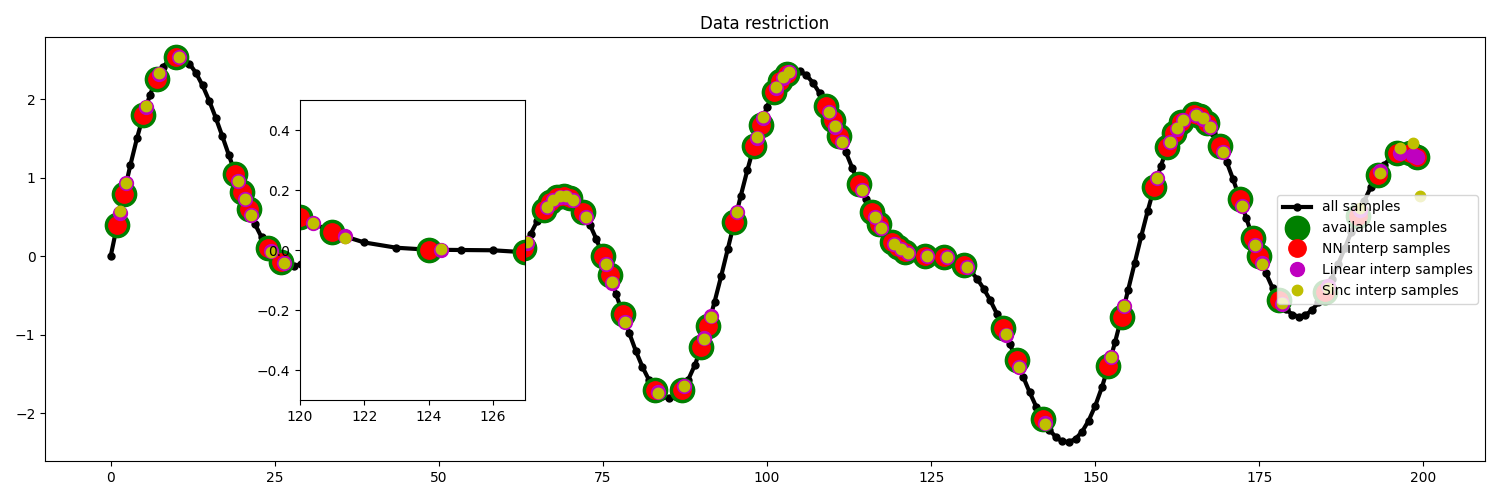
# Visualize data
fig = plt.figure(figsize=(15, 5))
plt.plot(isample, x, ".-k", lw=3, ms=10, label="all samples")
plt.plot(isample, ymask, ".g", ms=35, label="available samples")
plt.plot(iavann, ynn, ".r", ms=25, label="NN interp samples")
plt.plot(iavali, yli, ".m", ms=20, label="Linear interp samples")
plt.plot(iavasi, ysi, ".y", ms=15, label="Sinc interp samples")
plt.legend(loc="right")
plt.title("Data restriction")
subax = fig.add_axes([0.2, 0.2, 0.15, 0.6])
subax.plot(isample, x, ".-k", lw=3, ms=10)
subax.plot(isample, ymask, ".g", ms=35)
subax.plot(iavann, ynn, ".r", ms=25)
subax.plot(iavali, yli, ".m", ms=20)
subax.plot(iavasi, ysi, ".y", ms=15)
subax.set_xlim([120, 127])
subax.set_ylim([-0.5, 0.5])
plt.tight_layout()
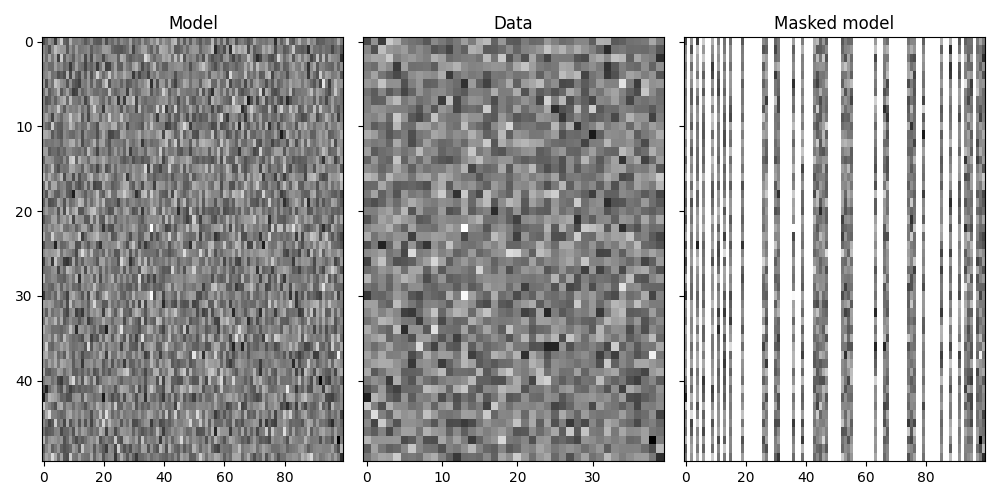
Finally we show how the pylops.Restriction is not limited to
one dimensional signals but can be applied to sample locations of a specific
axis of a multi-dimensional array.
subsampling locations
nx, nt = 100, 50
x = np.random.normal(0, 1, (nx, nt))
perc_subsampling = 0.4
nxsub = int(np.round(nx * perc_subsampling))
iava = np.sort(np.random.permutation(np.arange(nx))[:nxsub])
Rop = pylops.Restriction((nx, nt), iava, axis=0, dtype="float64")
y = Rop * x
ymask = Rop.mask(x)
fig, axs = plt.subplots(1, 3, figsize=(10, 5), sharey=True)
axs[0].imshow(x.T, cmap="gray")
axs[0].set_title("Model")
axs[0].axis("tight")
axs[1].imshow(y.T, cmap="gray")
axs[1].set_title("Data")
axs[1].axis("tight")
axs[2].imshow(ymask.T, cmap="gray")
axs[2].set_title("Masked model")
axs[2].axis("tight")
plt.tight_layout()
Total running time of the script: (0 minutes 0.759 seconds)